Ever since I first started learning about regression analysis, I found myself wishing I could do something equivalent to inspecting residuals for logistic regressions like you could with OLS. Earlier this week, I was looking around for more ways to spot-check logistic regression models, and I came across a plotting technique that’s described in this paper (Greenhill, Ward, & Sacks, 2011). They’re called “separation plots”, and they’re used to help assess the fit adequacy of a model that has a binary variable as its dependent variable. (Edit: ugh, I was on university wifi when I was reading and drafted out this post. I didn’t realize the paper was paywalled…now I can’t even reread one of the sections I wanted to quote!)
The authors did create an R package with a separationplot() function,
but I found that it was handling R’s graphic devices a little strangely,
so I reimplemented a basic version with ggplot2. Anyways, here’s
an example plot.
library(tidyverse)
library(scico)
fit1 <- glm(inmetro ~ percollege, data = midwest, family = binomial)
midwest$f1 <- predict(fit1, midwest, type = "response")
separation_plot(pred = midwest$f1, actual = midwest$inmetro)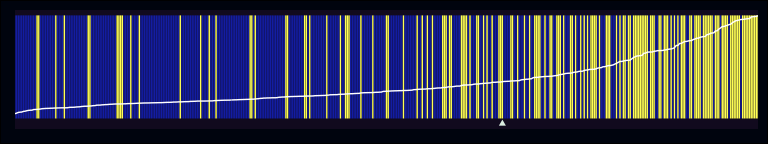
Using the midwest dataset from ggplot2, we’ve built an
underspecified model that’s meant to predict whether each county (from 5
midwestern states) is in a metropolitan area, based on the percentage of
adults that are college educated. So, what’s going on in this plot? Each
vertical line in the figure represents one of the 437 counties, with the
yellow lines reflecting events (i.e. instances where inmetro == 1).
Each vertical line in the figure is ranked in ascending order based on
the predicted probability output by the model. The further a vertical
line is to the left, the smaller its estimated probability.
A good model should be able to separate events from non-events, which should translate into vertical lines of the same color clustering together on their respective poles of the plot. For example, let’s look at an improved version of the model we fit earlier.
fit2 <- glm(
inmetro ~ percblack + percwhite + percollege + percpovertyknown + percbelowpoverty + percprof,
data = midwest,
family = binomial
)
midwest$f2 <- predict(fit2, midwest, type = "response")
separation_plot(midwest$f2, midwest$inmetro)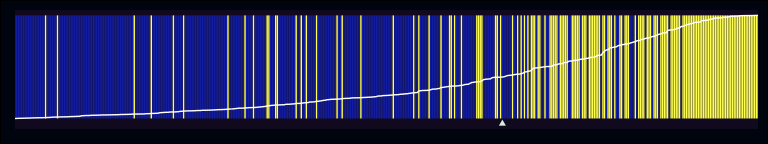
In comparison with the first plot, you can see that the non-events on the left are still being broken up occasionally, but these patches are less frequent and smaller. Conversely, on the right it seems like the second model is assigning higher probabilities to counties that really are in metropolitan areas. Let’s also talk about two other pieces of information that are being included in these plots. First, there’s the rising white line that runs the length of the figure. This simply represents the predicted probability of each observation in the model (with 0 at the bottom of the graph, and 1 at the top). As can be seen in the second figure, the slope of the line begins to rise more quickly in relation to the higher density of actual events. Second, there’s the little white triangle just below the lower edge of the plot. This is simply the sum of all the predicted probabilities, and I think is meant to serve as an estimate of the total # of expected events predicted by the model.
In the case of a perfect model, the marker should be exactly at the left-most edge of the events, like in the figure below.
# build a model that perfectly predicts each case
fit3 <- glm(inmetro ~ category, data = midwest, family = binomial)
midwest$f3 <- predict(fit3, midwest, type = "response")
separation_plot(midwest$f3, midwest$inmetro)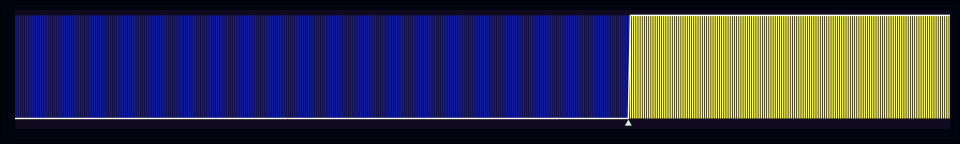
One of the things that feels appealing about this plotting approach is that you can see all the data that’s being modeled. Similar to residual plots, you’re able to get a sense of where fitted values are lying relative to the observed result. It seems like these figures could be an alternative or complement to ROC plots, and I’m curious if other folks find them interesting or useful.
Lastly, here’s the function if you wanted to roll your own ggplot.
separation_plot <- function(pred, actual, show_legend = FALSE, predline = TRUE, show_expected = TRUE) {
dat <- tibble(pred = pred, actual = actual) %>%
arrange(pred) %>%
mutate(position = 1:n())
p <- dat %>%
ggplot(aes(x = position, xend = position, y = 0, yend = 1, color = factor(actual))) +
geom_segment() +
scale_color_scico_d(palette = "imola") +
scale_x_continuous(expand = c(0, 0)) +
theme(
axis.text = element_blank(),
axis.title = element_blank(),
panel.grid = element_blank(),
)
if (!show_legend) {
p <- p + theme(legend.position = "none")
}
if (predline) {
p <- p + geom_line(aes(x = position, y = pred))
}
if (show_expected) {
p <- p +
annotate(geom = "point", x = max(dat$position) - sum(dat$pred), y = -0.05, shape = 17)
}
p
}